I did some research into this, and it looks like the fundamental reason for halos and lack of clarity in focus stacking is due to the fact that focus stacking software can't really tell a bright featureless fragment of your subject in focus from an out-of-focus halo of a bright speckle which is seen in the same direction but which is not in focus. One could try to design a 2D PSF (point spread function) which would do a reasonable job removing the halo - but only for a particular halo. E.g., PSF for a halo seen against dark OOF background will be very different from the case when the halo is against a sharp fragment of the macro subject.
And the ultimate fix for that (and also for increasing sharpness of diffraction limited image stacks) seems to be a blind deconvolution carried out in 3D (that is, applied to the stack, before focus stacking procedure). I tried (and failed) to find a readily available software for this purpose, either open source or commercial. There is one open source project which seems to be promising (AIDA - adaptive image deconvolution algorithm; https://github.com/erikhom/aida) - but the software seems to have been abandoned a few years ago, and the comments left there by people trying to use it are not encouraging (like, "it dosen't work with 3D stacks"). I tried to install it but failed, because I couldn't install the dependency (package Priithon - apparently another case of abandonware).
There is apparently an active ongoing academic research on this, in biological microscopy field. This is typically described as "blind deconvolution of Z-stacks in wide-field microscopy". One of the more recent and interesting papers on this topic is the following one:
Blind Deconvolution of Widefield Fluorescence Microscopic Data by Regularization of the Optical Transfer Function (OTF)
They claim their algorithm is much less prone to ringing artifacts - the most typical issue with any deconvolution of photographs. It also handles properly the noise in data.
Here is one of their tests:
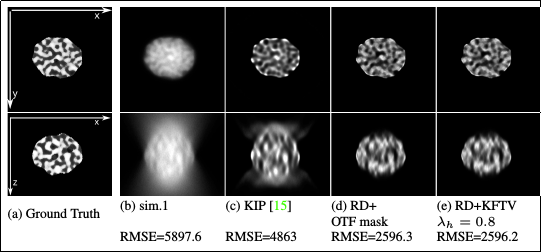
This is two slices through 3D focus stacks: top row is the XY slice, and bottom row is the XZ slice (Z being the optical axis of the microscope). The left images are the "true" ones (the model); the ones next to them to the right are for the original focus stack, with lots of OOF blur polluting both the image plane (XY) and the stack (XZ plane). It is obvious that if one applies focus stacking on this raw stack, it will be full of halos and will lack clarity.
Their case (c) shows a previous 3D blind deconvolution method applied to the 3D stack. It already helps quite a bit by removing bright OOF halos. But the (d) and (e) images (the proposed deconvolution algorithm) does a much better job, effectively getting rid of OOF light. These images would be perfect for focus stacking processing, as only sharp features are left in the 3D stack, so no confusion for the stacking software.
This all looks good in theory. The important questions are
- Will it work well for real life extreme macro focus stacks, and
- How hard will it be to write software to implement this method for macro photography.
I will try to write the software, but it will take a while.
Has anyone here tried 3D deconvolution for focus stacks? What was your experience with it?
